Basic Skills in Computational Research Environments
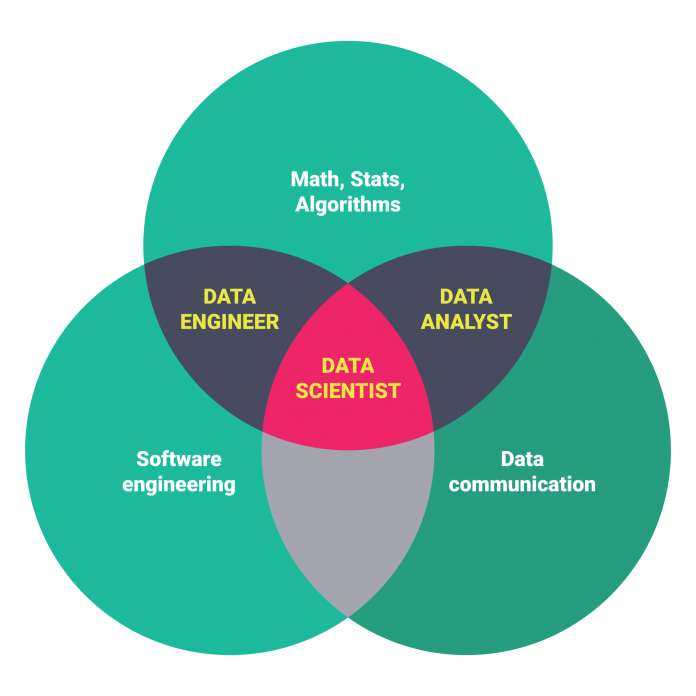
An early blog post about differences between Data Analysts, Data Scientists, and Data Engineers
Introduction
Big data Growth in data was foretold is a meme
Growth in data was foretold is a meme maybe_there_are_no_solutions.jpg, but different flavors of bioinformaticians can be referred to as analysts, data scientists, and data engineers. This article
maybe_there_are_no_solutions.jpg, but different flavors of bioinformaticians can be referred to as analysts, data scientists, and data engineers. This article no_shame_in_trying.jpg just brushes the surface of the differences and focuses the conversation on the reliance
no_shame_in_trying.jpg just brushes the surface of the differences and focuses the conversation on the reliance actually_a_brilliant_strat.jpg of individual team members on each other for hypotheses
actually_a_brilliant_strat.jpg of individual team members on each other for hypotheses We're made of star stuff. We're the way for the cosmos to know itself..., databasesIs MySQL Web Scale?, models
We're made of star stuff. We're the way for the cosmos to know itself..., databasesIs MySQL Web Scale?, models no_rly_how_many_people_can_do_that.jpg, calculations
no_rly_how_many_people_can_do_that.jpg, calculations muh_calculation.png, websites
muh_calculation.png, websites quality_hypertext.gif, reports
quality_hypertext.gif, reports she_went_and_told_Gladys.gif, and more. When I was in school, it was almost a joke that bioinformaticians who provide alignment, differential expression analyses, and modeling of transcriptomic data often get excluded
she_went_and_told_Gladys.gif, and more. When I was in school, it was almost a joke that bioinformaticians who provide alignment, differential expression analyses, and modeling of transcriptomic data often get excluded all_i_wanted_was_a_gd_stapler.gif from papers
all_i_wanted_was_a_gd_stapler.gif from papers 100% meritocracy in favor of scientists doing the laboratory work. I dont think the field is mature enough to understand the complexity
100% meritocracy in favor of scientists doing the laboratory work. I dont think the field is mature enough to understand the complexity imagination.jpg of computationxXxRoninC0d3r involved or how fortunate
imagination.jpg of computationxXxRoninC0d3r involved or how fortunate Number 1 reason: El Diablo burritos in PC it is that such work is even possible at this stage. By the end of the article, you should have a better understanding of the types of skills that can be useful, the strengths and weaknesses of certain flavors of bioinformaticians (data scientist vs data engineer), and how developing those skills at the expense of others will pidgeonhole
Number 1 reason: El Diablo burritos in PC it is that such work is even possible at this stage. By the end of the article, you should have a better understanding of the types of skills that can be useful, the strengths and weaknesses of certain flavors of bioinformaticians (data scientist vs data engineer), and how developing those skills at the expense of others will pidgeonhole quantitative_reasoning.jpg you into a role where it is your responsibility
quantitative_reasoning.jpg you into a role where it is your responsibility Wait... I'm my only advocate? to communicate value and preferences to your team. Bioinformaticians have some balance of software, comp sci, maths/stats, and scientific expertise
Wait... I'm my only advocate? to communicate value and preferences to your team. Bioinformaticians have some balance of software, comp sci, maths/stats, and scientific expertise
Data science People don't like statistics is not new. Data science
People don't like statistics is not new. Data science What's with the rebrand did not invent machine learning or modeling approaches any more than Six Sigma
What's with the rebrand did not invent machine learning or modeling approaches any more than Six Sigma Black belt btw invented statistics. Occassionally you hear something like 'Microsoft wants to solve biology' or 'AI experts claim they can out-compete the pharma industry' in headlines and it's just... cute. And I'm not even an expert.
Black belt btw invented statistics. Occassionally you hear something like 'Microsoft wants to solve biology' or 'AI experts claim they can out-compete the pharma industry' in headlines and it's just... cute. And I'm not even an expert.
A bit about my background
While studying biochemistry I learned some basic analytical chemistry and laboratory methods, but I wanted to move towards biology. I was discouraged at the pace of single-gene analyses in molecular biology given the scope of the systems involved and the degree of heterogeneity in cancer biology. After graduating I joined the Bioinformatics and Computational Biology program at my uni and moved into sequencing data analysis and associated work and I got a few second authorships under the guidance of K and T. The projects were enjoyable and the environment stimulating if not a little disheartening during the anti-science spending policies of the time, but research was still happening, and I was lucky enough to work on microbial transcriptomics data. Eventually I joined a research pharmaceutical firm where I worked until early May of 2019, a chance to practice and improve my software development and see how large the systems facilitating that level of research can really be. Many scientists avoid infrastructure work, sadly. The systems at play in those companies can be very complicated and web technology is a must when it comes to informatics in pharma. Right before I left I developed a talent for benchmarking and modeling the performance of my software which helps me as an individual better communicate value to teams. Okay, thanks for listening. Now let's talk a bit about skillsets and standards for industry bioinformaticians.
Analysts, Data Scientists, and Data Engineers
Every scientist has a flavor Taste all teh flavors and a role. Bench rats
Taste all teh flavors and a role. Bench rats cheesy_joke.jpg, desk jockeys
cheesy_joke.jpg, desk jockeys amirite_fellow_organics.gif, engineers
amirite_fellow_organics.gif, engineers good_times.jpg, and more. And each scientist computational or otherwise makes a choice about laboratory techniques
good_times.jpg, and more. And each scientist computational or otherwise makes a choice about laboratory techniques Multiplex != better, statistical skills
Multiplex != better, statistical skills More details below, computational abilities
More details below, computational abilities More details below, infrastructure, and reliance on homegrown or third-party tools. In addition, the complexity of the biological systems under investigation requires advanced modeling and simulation techniques in contrast to simpler hypothesis testing. I would gather that most bioinformaticians fall somewhere within the following 3 categories, with strengths and obligations accordingly. As you learn skills or specialize in these directionsOdd. It rings true even more since the pandemic. "A new scapegoat for tough numbers...", try to understand the parts of your team that are in the minority.
More details below, infrastructure, and reliance on homegrown or third-party tools. In addition, the complexity of the biological systems under investigation requires advanced modeling and simulation techniques in contrast to simpler hypothesis testing. I would gather that most bioinformaticians fall somewhere within the following 3 categories, with strengths and obligations accordingly. As you learn skills or specialize in these directionsOdd. It rings true even more since the pandemic. "A new scapegoat for tough numbers...", try to understand the parts of your team that are in the minority.
Scientists and Analysts
In my experience, most scientists can do basic regression FTW actually... and hypothesis
FTW actually... and hypothesis negative_results_are_still_results.png testing in Excel. Armed with educational or professional backgrounds in specialized biological or chemical knowledge, they have good understanding of experimental design
negative_results_are_still_results.png testing in Excel. Armed with educational or professional backgrounds in specialized biological or chemical knowledge, they have good understanding of experimental design Douglas C. Montgomery, Gil. Strang, CRC Press McElreath..., and scientific communication
Douglas C. Montgomery, Gil. Strang, CRC Press McElreath..., and scientific communication Is this a 'meta'?. However, their weakness in the computational side
Is this a 'meta'?. However, their weakness in the computational side Thought they'd be friends... of the field means they have little understanding of novel metrics
Thought they'd be friends... of the field means they have little understanding of novel metrics Foot_pound.png, modeling techniques
Foot_pound.png, modeling techniques Pragmatism > theory, or the costs
Pragmatism > theory, or the costs What about ethics?? Rust foundation? PyPI? EFF and FSF? associated with software design and calculations
What about ethics?? Rust foundation? PyPI? EFF and FSF? associated with software design and calculations Money_through_obfuscation.webp. One of the benefits of calculations, models, and simulation is the reduced cost of laboratory experimentation through prediction or simulation of results and scenarios that would require considerable experimentation to do otherwise. They are heavily reliant on graphical interfaces, websites, and databases. And they rely heavily on the following groups to build tools, critique third-party infrastructure, clean datasets, or provide models that can be used.
Money_through_obfuscation.webp. One of the benefits of calculations, models, and simulation is the reduced cost of laboratory experimentation through prediction or simulation of results and scenarios that would require considerable experimentation to do otherwise. They are heavily reliant on graphical interfaces, websites, and databases. And they rely heavily on the following groups to build tools, critique third-party infrastructure, clean datasets, or provide models that can be used.
Data Scientists
There are often more insights Rebranding_continuted.jpg to gain from existing datasets and not enough time or personnel
Rebranding_continuted.jpg to gain from existing datasets and not enough time or personnel This is why we can't have nice things... to model or suggest future sampling required for improved 'sensitivity'
This is why we can't have nice things... to model or suggest future sampling required for improved 'sensitivity' me_irl_irl.gif and specificity
me_irl_irl.gif and specificity the_most_secure_encryption.png of inferences. This has led to the rise of the data scientist specialization and new curricula designed to provide quantitative and computational skillsets to the next generation of analysts. With a strong background in matrix algebra and calculus, statistics, machine learning, and basic or intermediate software skillsets, data scientists are prepared to interpret larger datasets, perform calculations, and develop professional reports from combinations of new and old data.
the_most_secure_encryption.png of inferences. This has led to the rise of the data scientist specialization and new curricula designed to provide quantitative and computational skillsets to the next generation of analysts. With a strong background in matrix algebra and calculus, statistics, machine learning, and basic or intermediate software skillsets, data scientists are prepared to interpret larger datasets, perform calculations, and develop professional reports from combinations of new and old data.
They tend to have proficiency in R, Perl, Python, SQL, and shell languages. They are results oriented and make the most of existing infrastructure, not feeling shy to ask or query for datasets. However, their weakness in other pragmatic aspects of programming such as web development Basic_skills_means_scope_creep.webp, algorithm performance
Basic_skills_means_scope_creep.webp, algorithm performance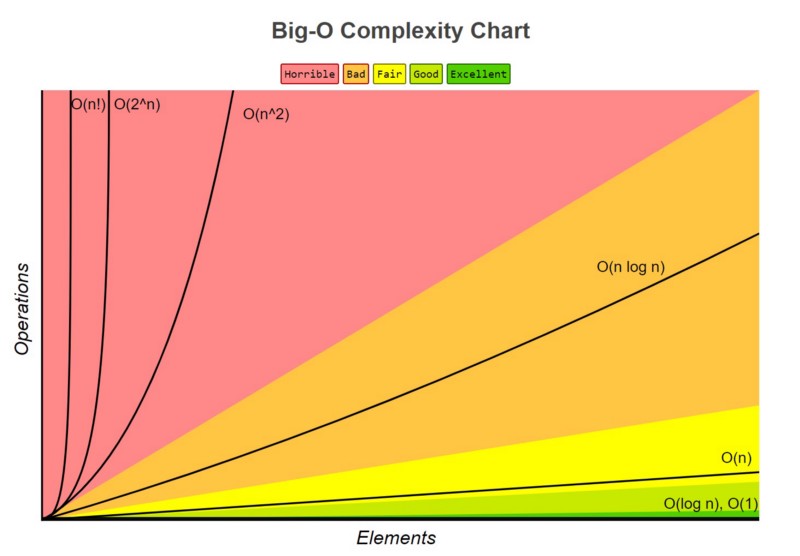 Scalability < sensibility, or systems design
Scalability < sensibility, or systems design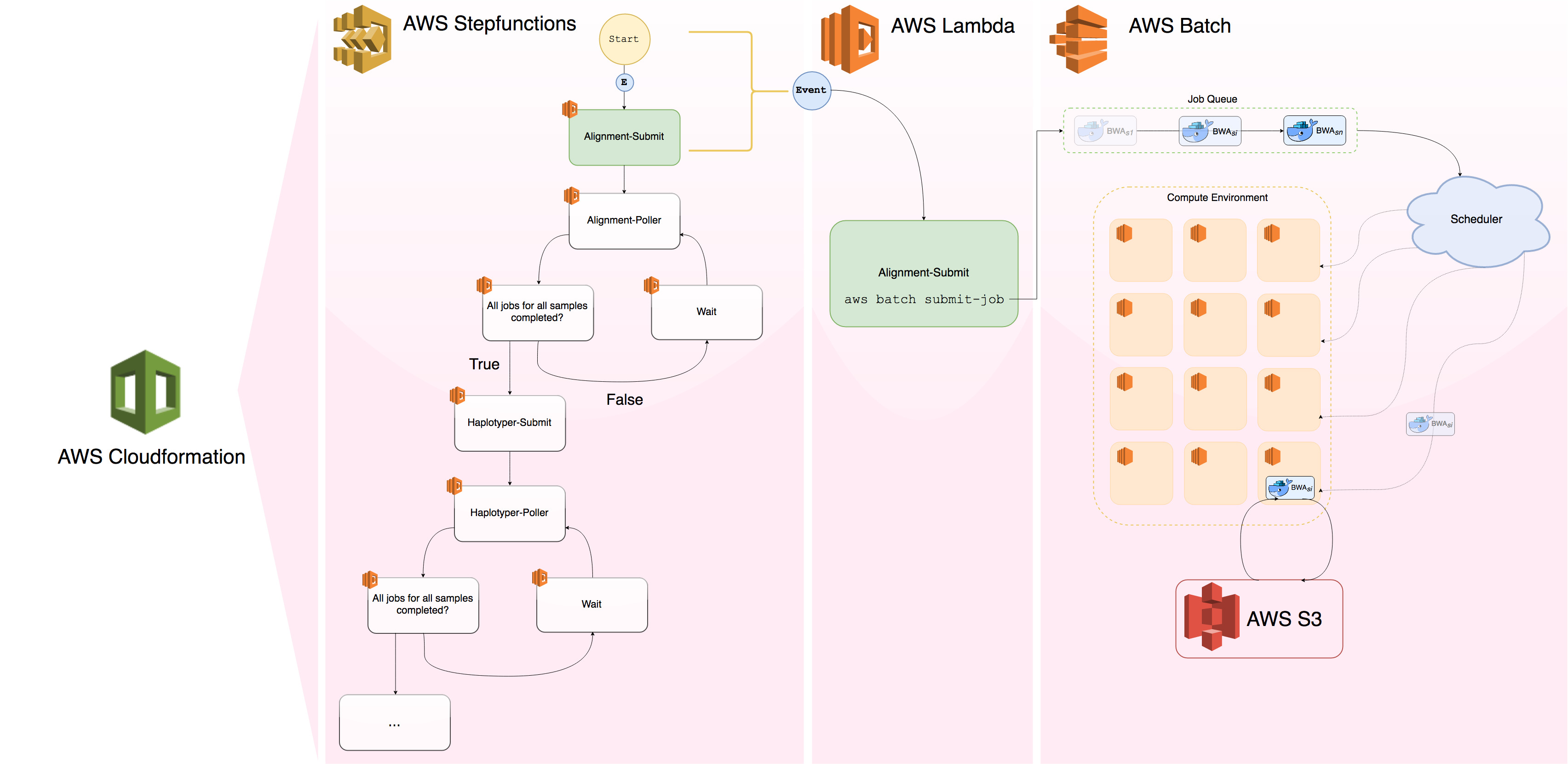 Basic haplotyper with scheduler is not emphasized by most data science curricula. Decisions are often made from their analyses and they have considerable decision making power. But again their job descriptions often focus on analyses and results instead of evaluating existing tools or building new infrastructure to enable others. They rely on existing databases and tools but juniors seldom participate in database or application designs, novel feature engineering problems (typically given to seniors), or improve the efficiency of existing tools.
Basic haplotyper with scheduler is not emphasized by most data science curricula. Decisions are often made from their analyses and they have considerable decision making power. But again their job descriptions often focus on analyses and results instead of evaluating existing tools or building new infrastructure to enable others. They rely on existing databases and tools but juniors seldom participate in database or application designs, novel feature engineering problems (typically given to seniors), or improve the efficiency of existing tools.
Data Engineer
The final category is also newer in its formalization, especially with the rise of larger datasets Bunnies and biology.. good reads actually..., databases
Bunnies and biology.. good reads actually..., databases Meme enjoyer, novice collector, or mixed numerical and document storage or abstract representations. Data engineers have rich software engineering and computer science abilities that lets them build critical infrastructure
Meme enjoyer, novice collector, or mixed numerical and document storage or abstract representations. Data engineers have rich software engineering and computer science abilities that lets them build critical infrastructure Not working? Dont fix it... for others with a focus on stability, usability, and efficiency. Pre-computating
Not working? Dont fix it... for others with a focus on stability, usability, and efficiency. Pre-computating What if we used 100% of our grad students? all the metrics, annotation
What if we used 100% of our grad students? all the metrics, annotation Not sure if there is metadata quality here... of datasets, and automation may be required for databases and applications in their wheelhouse. Enterprise software is often expensive to license and data engineers make a team responsive to the teams computational needs in a way that scientists and data scientists often cannot. However, I would say that these teammates, the data engineers, were an obvious minority in the teams Ive worked in. Their weakness is certainly a basic or non-specific background in the applied area and most companies do not support the growth of data engineers towards the scientific area. That said, they are invaluable teammates and can provide scripts, REST-APIs, databases, automated systems, web applications, and even some high-performance computingtest needs.
Not sure if there is metadata quality here... of datasets, and automation may be required for databases and applications in their wheelhouse. Enterprise software is often expensive to license and data engineers make a team responsive to the teams computational needs in a way that scientists and data scientists often cannot. However, I would say that these teammates, the data engineers, were an obvious minority in the teams Ive worked in. Their weakness is certainly a basic or non-specific background in the applied area and most companies do not support the growth of data engineers towards the scientific area. That said, they are invaluable teammates and can provide scripts, REST-APIs, databases, automated systems, web applications, and even some high-performance computingtest needs.
From basic computational skills to rather advanced, the needs of analytics in modern scientific fields cannot be overstated. Working in a team can be quite complicated and challenging, especially with the limited business, project management, and philosophical training that most S.T.E.M. curricula provide. Take for example a group I was in: the bulk of the deliverables were produced by theoretically focused scientists who were adept at running simulations in GUIs but only vaguely participated in infrastructure design, construction, or maintenance.
Producing deliverables to other groups is our most important role, from the perspective of those outside ours. However, the theoretical scientists efficiencies can be enhanced with the appropriate infrastructure or selection from a diverse range of modeling tools. The divide between the engineers and theoretical scientists is natural in these groups but reinforces boundaries between those with visibility to the rest of the company vs. those that enable the group through software and algorithm performance.
Difference between software dev and comp sci?
Software dev and project management
Software development or software engineering is a discipline that I would argue is very humanistic, highly in flux, and largely driven by ecommerce. Software engineers build calculators gimme_da_answer.png, databases
gimme_da_answer.png, databases Jesus Christ..., its Jason Bourne, applications
Jesus Christ..., its Jason Bourne, applications nah_but_scheme_was_tite.png, and software systems
nah_but_scheme_was_tite.png, and software systems x1337hax0r322 that enable other members of their organization. Software engineering tools focus on application frameworks
x1337hax0r322 that enable other members of their organization. Software engineering tools focus on application frameworks totally_modern_meme.webp, project management skills, and interface design to make the product as mature and useful
totally_modern_meme.webp, project management skills, and interface design to make the product as mature and useful hawaii_missile_alert.gif for the end-user while maintainable for other software developers. Software development skills include version control systems, or VCSHulter tries git. Arguably the most popular version control system at the moment is git
hawaii_missile_alert.gif for the end-user while maintainable for other software developers. Software development skills include version control systems, or VCSHulter tries git. Arguably the most popular version control system at the moment is git I prefer final_project_LAST_2019-01-22.tar.gz.
I prefer final_project_LAST_2019-01-22.tar.gz.
Other useful skills for software developers to have (and maintain) include software testing strategies like the so called Behavior Driven Development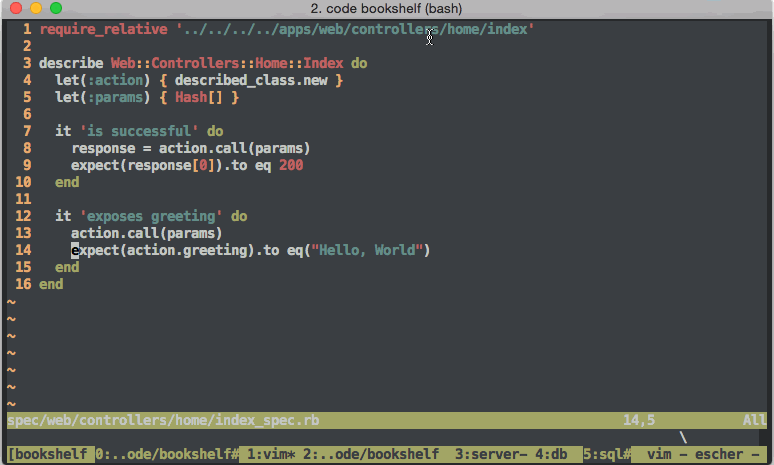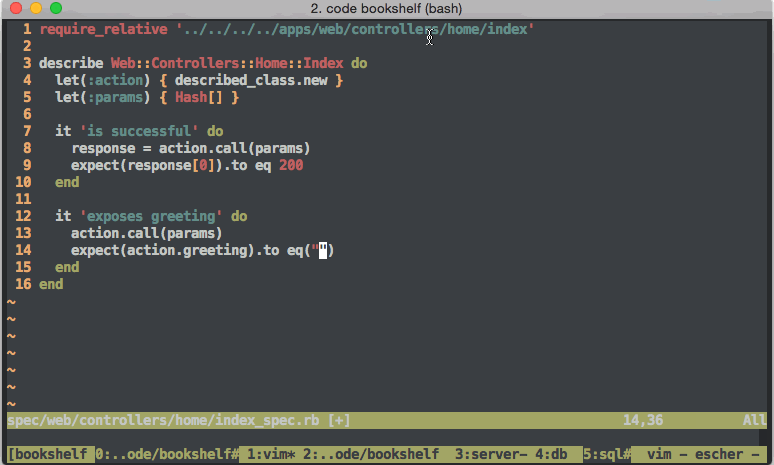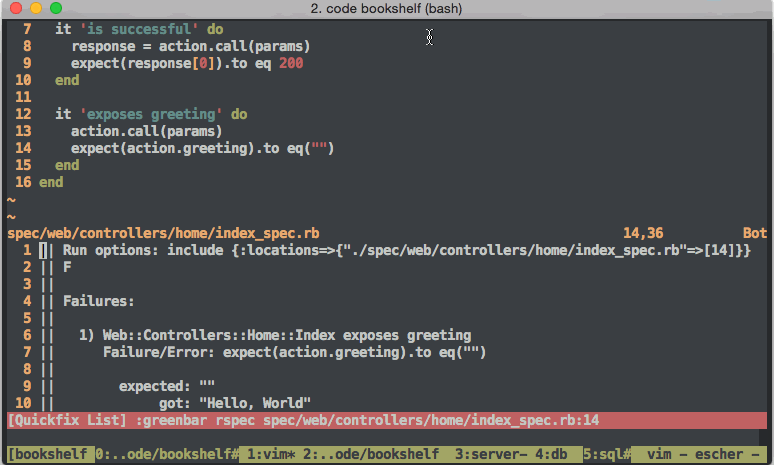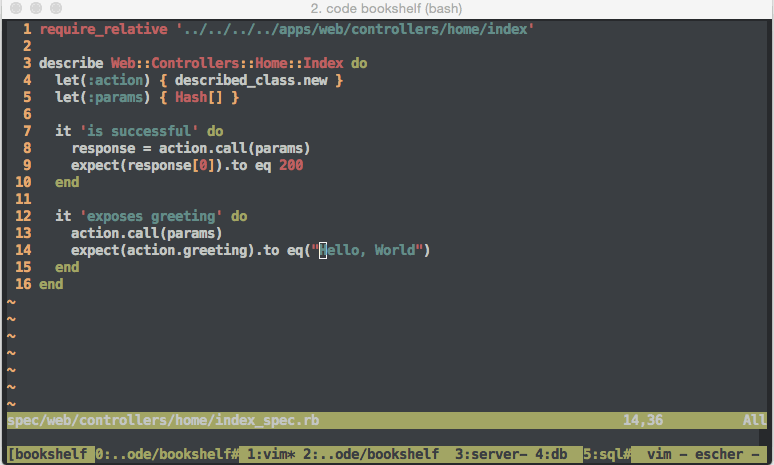 If you want.. or BDD, and its older cousin Test Driven Development or TDDHoltre tries unit testing.. In fact, the whole world of software development has a new era feel ever since Web2 made large centralized software development include features of rapid iteration and testing. The world of DevOps isnt worth getting into. No seriously, dont do it. DevOps teams dont even do it. However, interesting features for larger software development organizations include practices such as continuous integration (CI)
If you want.. or BDD, and its older cousin Test Driven Development or TDDHoltre tries unit testing.. In fact, the whole world of software development has a new era feel ever since Web2 made large centralized software development include features of rapid iteration and testing. The world of DevOps isnt worth getting into. No seriously, dont do it. DevOps teams dont even do it. However, interesting features for larger software development organizations include practices such as continuous integration (CI) halfway_awful_devops_graphic.jpg, package management
halfway_awful_devops_graphic.jpg, package management shameless_technical_opinion.png, web design
shameless_technical_opinion.png, web design do_people_rly_enjoy_this_stuff.webp, and user interfaces or UI
do_people_rly_enjoy_this_stuff.webp, and user interfaces or UI okay_not_as_easy_as_i_thought.gif.
okay_not_as_easy_as_i_thought.gif.
There is an entire rabbit-hole of software engineering skills to learn the terminology, consider the use-case, and drop it like a hot potato. Consider software systems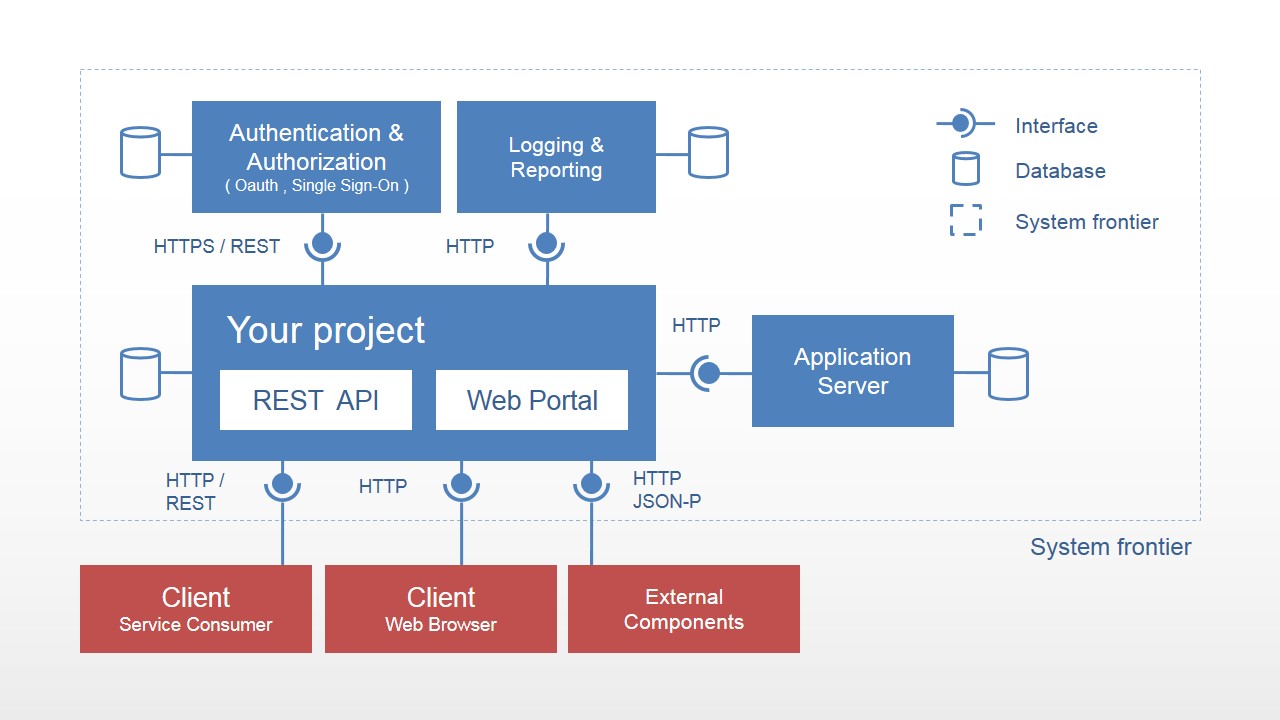 test architecture, automaton paradigms
test architecture, automaton paradigms something something bare metal, and iron and wine? for instance. When automating your paradigms, youll want to ensure your code is tested, because observing murphys law...you know that whatever can happen will happen, often for the worse. For this reason, faultyDungeon raid... but also.. this was long before Linus had issues with one of his employees. software will need to be redacted and entire studies may be removed, crippling a researchers career. Understanding application fault toleranceI need to touch grass.
allows us to build reliable applications that are more resilient to subsystem failures, logging the errors rather than crashing the server, and other system quality considerations.
something something bare metal, and iron and wine? for instance. When automating your paradigms, youll want to ensure your code is tested, because observing murphys law...you know that whatever can happen will happen, often for the worse. For this reason, faultyDungeon raid... but also.. this was long before Linus had issues with one of his employees. software will need to be redacted and entire studies may be removed, crippling a researchers career. Understanding application fault toleranceI need to touch grass.
allows us to build reliable applications that are more resilient to subsystem failures, logging the errors rather than crashing the server, and other system quality considerations.
But then again, on the simpler side... pre-deployment? Theres a ton of skills to master there too. User interface (UI Can it be that it was all so simple then?) is the tip of the ice berg when building products that colleagues, customers, professors, and other students will want to use.
Can it be that it was all so simple then?) is the tip of the ice berg when building products that colleagues, customers, professors, and other students will want to use.
Like many professions, time management and documentation are crucial skills for describing work that has been done or is required for meeting milestones, deadlines, and budgetary guidelines. Project management skills include documentation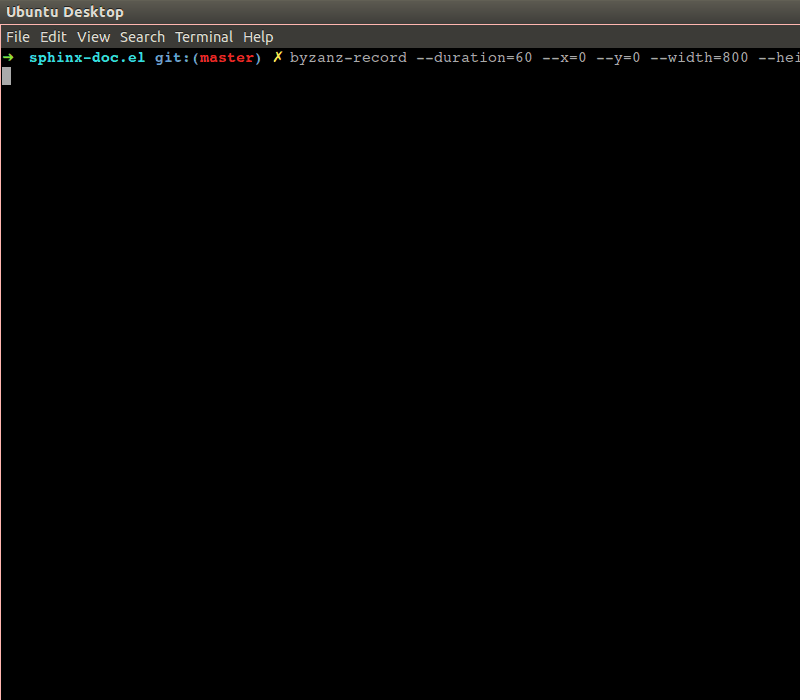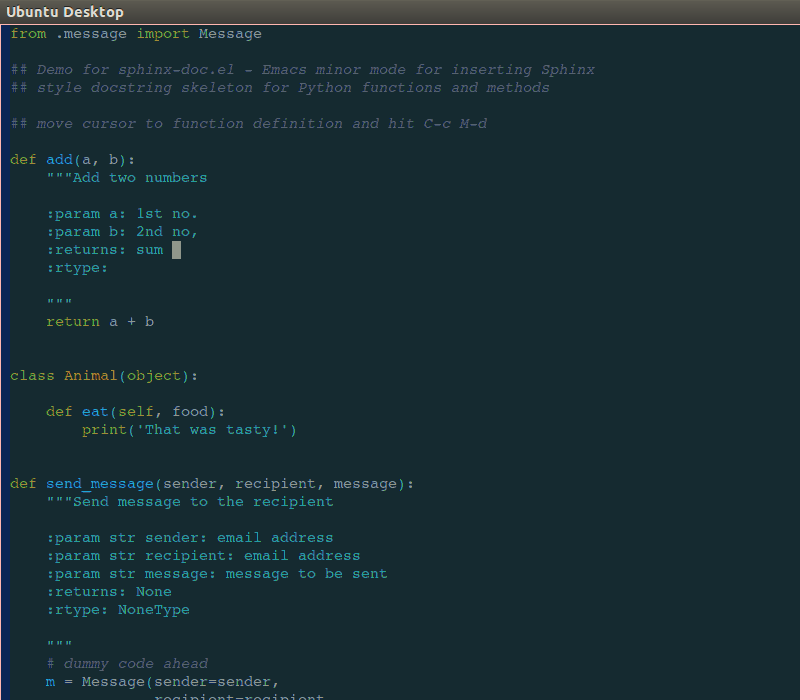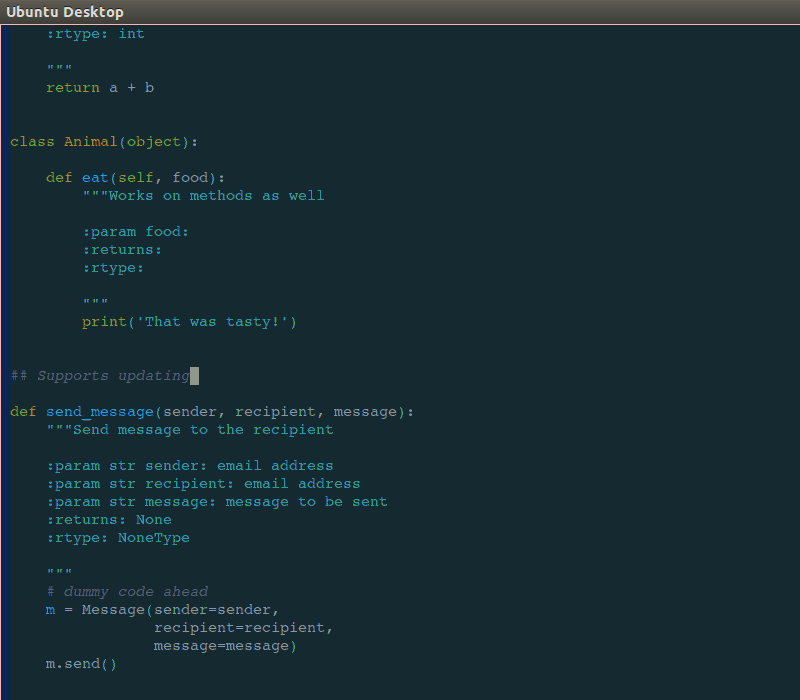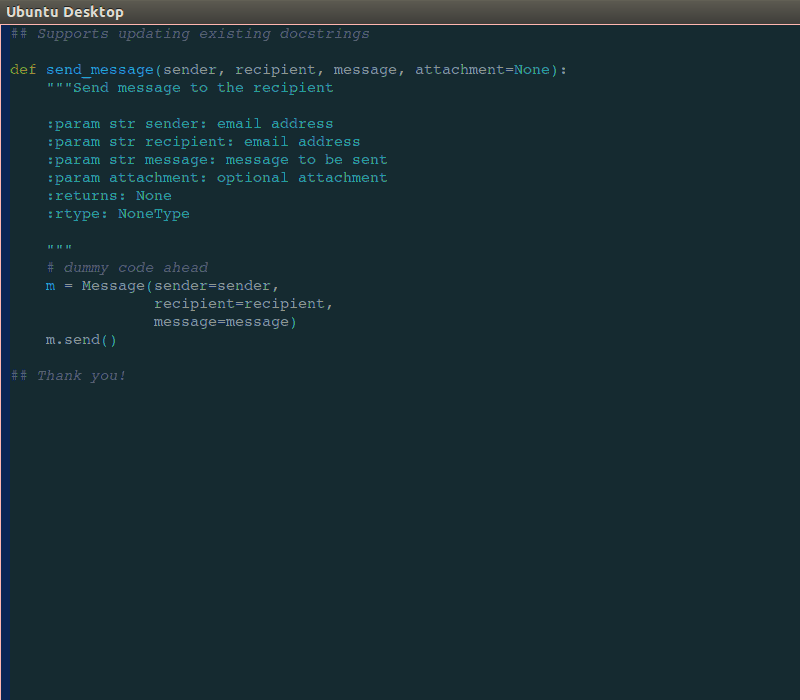 handy Sphinx docstrings into ReadTheDocs for Python code, kanban, Github, Atlassian Confluence and Jira, and more traditional Gant charting, minimum-viable product (MVP), budgeting, and cost projection skills.
handy Sphinx docstrings into ReadTheDocs for Python code, kanban, Github, Atlassian Confluence and Jira, and more traditional Gant charting, minimum-viable product (MVP), budgeting, and cost projection skills.
It definitely feels like project management courses should be part of every graduate engineering proram, even if the project-focus teaching style (problem sets, end-of-course projects) is present in addition to a thesis/dissertation. Theres no discussion of common business, budget, and timeline questions like "here's the questions employers will ask when you're overbudget or late," or "here's the things you should routinely do to communicate value for a project of this size," or "here's how you can recruit coworkers and delegate properly so that you don't become overwhelmed."
Computer science, math, and natural science
If software development is like the business side of bioinformatics... communicating value, timelines, budgets, stability, usability, maintainability... then computer science is a logical foundation that provides value to calculations, pipelines Did I miss the requirement to feel?, models, or applications. Computer science certainly emphasizes algorithmic performance (space
Did I miss the requirement to feel?, models, or applications. Computer science certainly emphasizes algorithmic performance (space space_issues_are_common_in_bioinf.svg and time
space_issues_are_common_in_bioinf.svg and time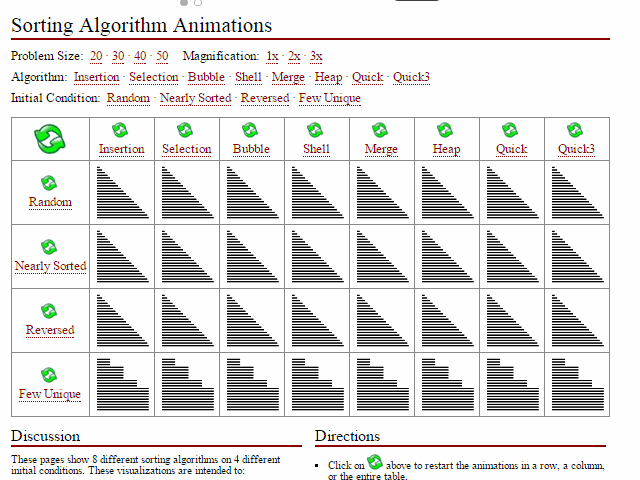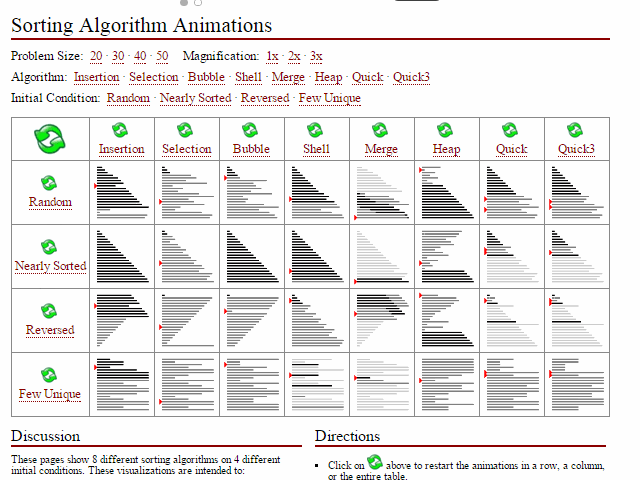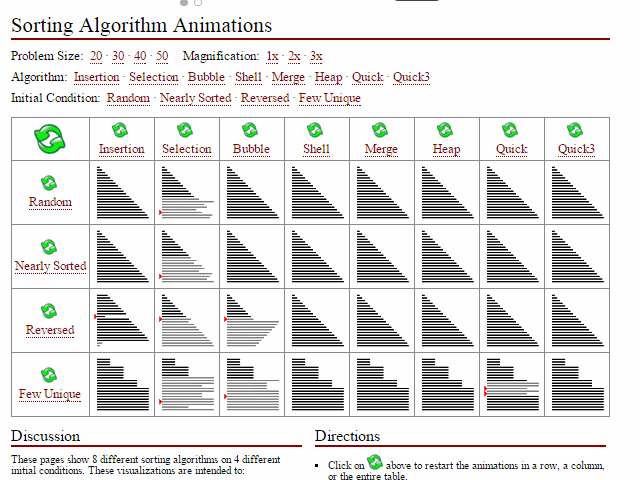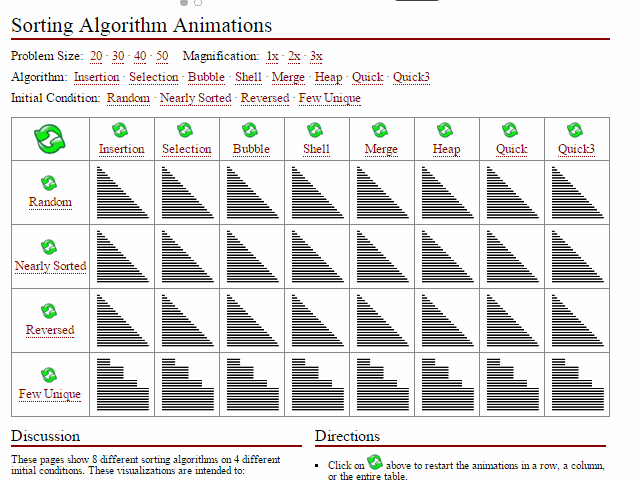 interesting_sort_comparisons.gif). I can' undersell that enough here; these are incredibly important first principles for compsci students including skills such as big-O notation, high-performance computing
interesting_sort_comparisons.gif). I can' undersell that enough here; these are incredibly important first principles for compsci students including skills such as big-O notation, high-performance computing ...test (HPC), compiled
...test (HPC), compiled flavors.jpg languages
flavors.jpg languages best_childhood.jpg, parallelization
best_childhood.jpg, parallelization always_a_bottleneck.jpg and concurrency
always_a_bottleneck.jpg and concurrency can_we_get_some_doggone_performance_gains.webp, floating point accuracy
can_we_get_some_doggone_performance_gains.webp, floating point accuracy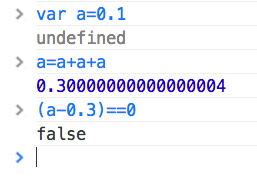 check out 'wat' bby Gary Bernhardt on YouTube, Occam's razor
check out 'wat' bby Gary Bernhardt on YouTube, Occam's razor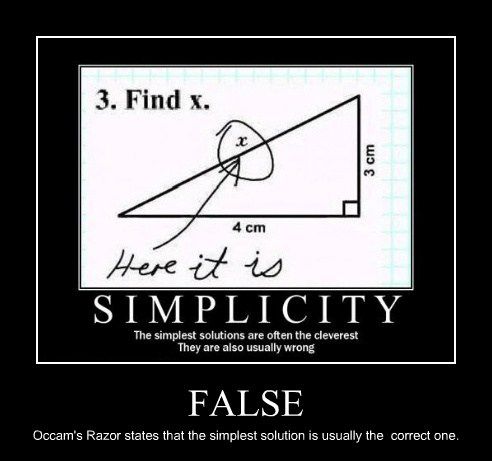 arguably_doing_it_right.png, and engineering assumptions.
arguably_doing_it_right.png, and engineering assumptions.
But the discipline has a wide variety of applied areas that work on various data types from primitive (float a decimal is a mantissa and an exponent, int
a decimal is a mantissa and an exponent, int an integer can have 8, 16, 32, or 64 bits of precision, string
an integer can have 8, 16, 32, or 64 bits of precision, string a list of ASCII characters. Words 'n stuff..., array
a list of ASCII characters. Words 'n stuff..., array Rebuttal, hashmap
Rebuttal, hashmap Big data hadoops in the cloud at web-scale) to complex (nested structures, document storage, object-class hierarchies, pickled objects), as well as data location (memory, cache, in-memory cache, cloud, object storage, database, filesystem, compression). Computer science and mathematics allows researchers to derive the right metric for a model to provide better predictive or inferential power. Interdisciplinary science education cannot be expected to adequately cover the diversity of programming paradigms, languages, and frameworks available nor the historical, current, and modern algorithms or modeling techniques used in the field. My program provided an overview of python, fasta parsing, alignment algorithms, web tools, and database technology to make room for advanced topics in biology. Most bioinformaticians-in-training could benefit from workshops on benchmarking, big-O complexity, parameter sweeps, and similar metrics that can help you quantify the value of your implementations.
Big data hadoops in the cloud at web-scale) to complex (nested structures, document storage, object-class hierarchies, pickled objects), as well as data location (memory, cache, in-memory cache, cloud, object storage, database, filesystem, compression). Computer science and mathematics allows researchers to derive the right metric for a model to provide better predictive or inferential power. Interdisciplinary science education cannot be expected to adequately cover the diversity of programming paradigms, languages, and frameworks available nor the historical, current, and modern algorithms or modeling techniques used in the field. My program provided an overview of python, fasta parsing, alignment algorithms, web tools, and database technology to make room for advanced topics in biology. Most bioinformaticians-in-training could benefit from workshops on benchmarking, big-O complexity, parameter sweeps, and similar metrics that can help you quantify the value of your implementations.
Skills recommended for a balanced skillset?
Again, this is not a curriculum or even a suggestion for a particular bioinformatics course's focus. But I would say that there are some considerations every bioinformatician makes when deciding which paradigm/language/framework to invest in and this is my best overview of fundamentals that are expected from entry-level bioinformatics developers.
Data formats, data structures, and databases
Almost all data that we work with is ASCII un gato, from simple .tsv
un gato, from simple .tsv ms paint expanding brain, fastasthx, good fun. and fastqs to abstract
ms paint expanding brain, fastasthx, good fun. and fastqs to abstract It's technically XML. and extensible formats like (a)cyclic network formats, PDBs, 2D/3D chemical structures, or tree structures. Knowlede of compression and encryption is also essential for working with clinical and compressed large datasets. For reference, the 1000 genomes project sequenced over 260 terabytes and the Broad Institute has sequenced over 70 petabytes compared with the size of the Netflix library of 3.14 petabytes. Other data furmats are emerging that are loosely constrained (JSON
It's technically XML. and extensible formats like (a)cyclic network formats, PDBs, 2D/3D chemical structures, or tree structures. Knowlede of compression and encryption is also essential for working with clinical and compressed large datasets. For reference, the 1000 genomes project sequenced over 260 terabytes and the Broad Institute has sequenced over 70 petabytes compared with the size of the Netflix library of 3.14 petabytes. Other data furmats are emerging that are loosely constrained (JSON meh, kind of cool) and are the basis for more complex data structures or databases. Validating and specifying complex structures is an ongoing problem and familiarity with json-schema is a plus. Or minus...
meh, kind of cool) and are the basis for more complex data structures or databases. Validating and specifying complex structures is an ongoing problem and familiarity with json-schema is a plus. Or minus...
Database technologies like MariaDB(formerly MySQL), PostgreSQL, and MongoDB are mainstays of enterprise data systems and web applications. A limited understanding of the entity-relationship model and normalizations isn't going to lose you the job, but it will make you a more effective web developer and data scientist.
But the most challenging aspect of modern data-driven fields is the tradeoff between the minimal amount of data for a complete record and the maximum amount of flexibility to anticipate the community's uses. I'll use the example of 2D/3D chemical structures to illustrate.
SMILES is a landmark cheminformatics format, made in the 1980s, for representing the network of a molecule's atoms in an easy-to-read format, superseded recently by the less easy-to-read IUPAC format InChI with better normalization and querying capabilities. The alternatives were hard for humans to read without the aid of GUIs, poorly extensible, and had a maximum record size of 999 atoms due to the Fortran record format of the original implementation. This format is still relevant today because of how easy it is for computational scientists to read or quickly understand what a molecule's composition is at the command-line. Fasta is a similarly simple format but is almost impossible to understand by reading long and low-dimensional nucleotide sequences.
Crystallographers required a larger and more extensible format for larger chemical structures and biomolecules and thus the PDB format was born. The modern PDB format is not human readable and requires the aid of a GUI to visualize but it supports metadata and other extensions. But with all of the available formats for cheminformatics, one has to wonder why efforts to parse and perform calculations on chemical formats are so disjointed, adding to the issue the near-infinite scope of chemical structures, macromolecules, and issues of post-translational modification, chemical protection, and cleavage sites. In summary, many different ecosystems of formats, parsers, and convenience functions get created with sometimes different goals: easy-to-read at the command line vs flexible and extensible formats for more complex data structures. This dichotomy is very relevant for bioinformatics.
So what makes a file format and supporting software 'good'? For example, BAM isn't particularly earth-shattering in its computational basis. It's a tsv format with a compression layer that shows alignment, sequence, quality, and mapping statistics, generated from much simpler fastq files. The implementations Picard and SAMtools are both high-quality with differing opinions on single-record and whole-sample validity. In my opinion, it's a really good implementation from an efficiency and usability perspective, with standard support for compression and indexing, a good test suite, and community support. What is amazing about it is the quality of the specification. As a result there is excellent adoption rate, and consistent implementation.
Data formats are essential for bioinformaticians, but many formats have limited community support, are redundant with other formats, or don't anticipate the community's needs. And most bioinformatics curricula do not include a critique or survey of existing formats, ideas for consolidation, or discussion of differences in formats as essential for NGS technology as SAM/BAM. These are skills and perspectives that experiencetest can remedy, but the nature of those formats and the depence of software systems on their consistency should be taken in to account when building 'futureproof' systems of automated processing, warehousing, annotation, and even analysis. All of those systems are then built with languages and programming paradigms that are also evolving and need consideration during each bioinformatician's training, and advice on these subjects is typically neglected.
Languages, paradigms, and problem solving
I'd like to leave the topic of data formats and databases to talk for a moment about the programming languages that we use to build our tools and systems, and the paradigms that influence our problem solving strategies. Many bioinformaticians are first exposed to easy-to-use interpreted languages like Python or Perl, with little attention given to shell proficiency, operating system tools, compiled languages, and the nature of debugging Types, Enums, Function signatures, Errors, Results, Options... single tools or whole software systems. Advanced programming isn't taught often to graduate bioinf. students and novices make numerous choices novices without much top-down support about why conventions, dichotomies, or pluralities exist.
Types, Enums, Function signatures, Errors, Results, Options... single tools or whole software systems. Advanced programming isn't taught often to graduate bioinf. students and novices make numerous choices novices without much top-down support about why conventions, dichotomies, or pluralities exist.
These days, memory issues are about as common as computational efficiency with the datasets we work with and can't be solved by simply switching to the steeper learning curve but better performance of compiled languages. Consequently, we often throw compute power and parallelization at most scientific tasks instead of switching to compiled languages. Type-checking and format validation is almost an afterthought for beginner bioinformaticians but is the most common errors beginners face. Rather than make a suggestion here, I'll just say that I've tried to balance my dialects with Python, R, shell, Ruby, and some novice level Haskell. I'm watching the Julia project with anticipation, but I doubt that I'll find much acceptance in industry. But in summary, static typing and type-checking is often a better solution than using a purely dynamic coding style or using more verbose compiled languages.
I'd claim from my experience that object-oriented programming isn't a bona-fide paradigm but instead is a construct for complex data-structures and the resulting encapsulation (part of the definition) with some aspects of declarative or imperative thrown in. I would suggest that most new bioinformaticians look at functional programming and type-classes instead of learning object-oriented programming in depth, trying not to glaze over at the abstract algebra involved to understand the merits of compiled languages and static typing, and how scientific such languages can be. Eliminating type errors reduces debug time, iteration time, and gets developers to the qualitative issues of their software much faster.
Every bioinformatician needs to know two paradigms: functional/procedural and event-loop. I'll go out on a limb and suggest that functional and procedural programming are simply distant cousins and should be treated as one overarching paradigm. Functional implementations are easy to understand with our mathematical backgrounds. Sometimes procedural implementations and familiarity with language internals/optimization makes code run faster but look less pleasing to understand. The only other programming paradigm in my understanding is event-loop languages like Javascript or Erlang. Concurrency can lead to considerable speedup in web applications or other systems that rely on external networked calls, but Javascript is a language of user-interfaces and web development and can be essential for data scientists looking to present tables, interactive graphics, or mature interfaces to their users.
The language ecosystem is the final under-described consideration that informs beginners and novices of language choice and influences intermediate users to switch to modern, marketable, and maintainable tools. Package management and security are important factors for enterprise systems, but more important are testing and documentation standards or heuristics/shortcuts taken by standard library algorithms. You'll know what I mean when you encounter a shortcut in the standard library that is not correct, but correct 'enough' for web developers and ecommerce. In summary, choose a language with a strong standard library, documentation, conventions for package structure and installation process, and with a strong bioinformatics ecosystem.
Rather than focus on problem solving paradigms directly, I spent the time talking about type-classes, two major programming paradigms, and ecosystem considerations when you choose which languages to develop skills with. Each problem has multiple approaches, where parallelization and concurrency can dramatically effect system/calculation performance. Some problems have redundant calculation, or frequently 'visited' substructures in a larger database, format, etc. where cache or index support should be provided. These issues affect both larger software systems with elaborate graphical user interfaces (GUIs) and simple command-line tools (I like to call them calculators) that are mainstays of the bioinformatics community.
User interfaces
I'll focus next on two types of user interfaces that developers should be familiar with consuming or producing for their customers, collaborators, end-users, or organization. The first thing to consume or build is of course the command-line interface. I'll describe an opinion here about an ideal Python-based calculator with subcommands. It should have no suffix (mycommand instead of mycommand.py) and a shebang should be present in the entry point. It should use STDIN/STDOUT preferably over --infile or similar. It should make use of standard argument parsing libraries like argparse where possible to make the code maintainable and extensable and built in support for -h, --help on the base command and all subcommands. git and the aws-cli are good examples of mature hierarchial utility command interfaces. A Makefile is a great entry point for auto-documentation, installation, and testing.
If your project is Pythonic, your package should have both a 'requirements.txt' and a 'setup.py' (which could reference the former) to facilitate the installation into the appropriate locations in the user's environment. Tests should be written (famous last words, I know) to show the stability of the code to different function inputs, expected errors, and interactions between modules/classes. Acceptance tests should be written to show that the CLI responds to input in the right way and produces correct calculations, conversions, or other outputs under various (un)expected situations. Test datasets aren't always easy to think of, but your code should handle good and bad quality data as you expect, not throw cryptic errors that reflect what you consider to be invalid. Finally, your repository should follow good git practices and you should branch, rebase, and squash according to what most would consider to be best practices. And to top it off, you should let a continuous integration server like Jenkins or Travis-CI run your tests for you! This is just my opinion of what a usable tool could look like to get good acceptance and support from the users.
But in addition, I think that most command-line tools should also have a API/SDK that makes functions/classes available to users who want to integrate it into their codebase. System calls are ugly. It looks cleaner and it's usually easier to test if you spin off the logic of your calculations into modules/classes and then call these as needed for your subcommand. No one wants to maintain or debug a 2k+ line script with a monolithic 'main' function. It's very easy to have a module directory, 'bin', 'docs', 'test', and 'test/data' directories to make a complete CLI tool.
And then there is the REST-API. REST-APIs are mainstays of modern software system architectures and web applications and ultimately provide the developer with a larger audience. They can contain the logic of a web application that can be used by scientists who might not be comfortable at the command-line, by data scientists who want broad access to available data, or for command-line interfaces that send/receive messages or data to the database. LDAP and authentication can come in to play for enterprise systems. 500 response codes are unacceptable: either the request was invalid or the server didn't find/respond in a way that produced a result... but something should always be returned, even an empty data structure. There are plenty of HTTP response codes that can tell the web page, CLI, or other component of the system why the request didn't produce a result. Many people like to use a reponse JSON object that looks roughly like the structure below. I'm still a beginner/intermediate software developer and some software systems can be rather complex, but an understanding of REST-APIs is a good foundation for bioinformaticians seeking to access databases without server-side access, to expose their calculators to a broader audience, or to develop software systems that have considerable value for their organization or audience.
{
success: True,
data: [...],
message: "this tells you what was done or what errors were produced"
}
Interface is a consequence of any calculation that you find useful and you'll find some you like, some you don't, and a whole bunch in between. Rather than describe any javascript or MVC framework you should go learn, I just decided to keep it non-commital and say that the choice of interface is important when you're deciding the scope of a project and timelines. Your interests in UI will adapt to the needs of your audience (e.g. a few dozen scientists) and the skills of your collaborators (e.g. half a dozen computational scientists).
DevOps, SysAdmin, and Linux fundamentals
The rise of Amazon's AWS platform has changed the game slightly for developers seeking speicalized hardware or burst capabilities. Developers are being put closer to budgets and efficiencies of their own software. In addition to Linux fundamentals (below), developers are often expected to have a basic understanding of the resources and cost savings available to them with cloud plaforms like Google, AWS, RedHat, and other cloud providers. I won't go in to extensive detail about the automation and systems tools available on those platforms at this stage, but I'd like to reinforce that the ecosystem and tools available to launch, update, and maintain software system components in different location has matured substantially and the traditional sysadmin roles are changing, as are expectations about top-tier developers.
Linux fundamentals
sed, awk, grepparallelcron, systemd timerssystemd, initd/dev, /etc, /mnt, $TMPDIRnfsPBS, SGE/UGE, torque, SLURMhttpd, nginxtmuxnvim
There are plenty of manuals and tutorials available for these topics so rather than reiterate here, skip to the next part if you have a basic understanding of what these tools do.
sed is a stream editor that edits each line of a piped stream or file according to a regular expression. Need to replace a string in your file/stream? Need to delete quotes? How about remove trailing whitespace or text from a column? These pretty common tasks can be done non-interactively as part of a shell-script or pipeline. If your dataset is too large to fit in memory and edit with emacs, nvim, or something similar, go ahead and use sed to perform subsequent edits using the power of regular expressions.
awk is an older stream processing language that can do remarkable things, similar to perl one-liners. However, our knowledge of scripting basics often makes it easier to create a one-off Python/Perl script that provides the same functionality and doesn't require learning a whole new language (it's technically Turing-complete). Occassionally you can't do a stream reformating operation with sed, need to skip lines or similar, rearrange columns or something and awk will be a better fit.
grep is another regular expression tool that can filter or locate lines in a file matching the expression. Filtering and finding text in a file is pretty common; search features are built into everything these days. Do it on the fly, non-interactive as a part of your pipeline or script, filter log files for certain warnings or errors. It's a pretty easy to use tool and is a great segue to sed
parallel is an optional tool in most Linux distros. It provides simple and performant parallelization of shell-functions, scripts, or simple commands. Want to build easy parallelization into your pipeline or processing? Use something like parallel 'program -k {1} -i {2} {3}' ::: $(seq 6 12) ::: $(seq 1000 5000) ::: $(ls /path/to/*.fastq.gz) for a parameter sweep over a range of input fastqs and parameter values (k, i). It's a great tool for production workflows and parameter sweeps during algorithm development, especially if it's too complicated to hardcode parallelization in your script, or you just want to do the same thing with different parameter values (a sweep) or across a number of input files.
cron is a tool for chronologically-based execution of certain programs (every day at 12am) and is a good tool for building software systems. Update a database, load new data, process that night's dataset, or create system backups. The primary command-line tool is called crontab and is included in most Linux distros. There is a specific columnar syntax to use when specifying when and how often a command should be run. Systemd timers are a modern replacement for the traditional Linux cron.
systemd is the replacement of the traditional process hierarchy in Linux (initd). This is an OS system that is launched early during the boot process to launch other daemons, mount hard drives, connect to the network, launch the firewall, etc. The original process launched in Linux has a process id (pid) = 1, and is usually owned by root. The init process can launch processes as other users for the session to use. systemd is a more advanced process management tool that would ultimately start your Apache httpd server and your web application or ensure automatic startup of those systems after system reboots. Some Linux servers might be on 24/7, might not be rebooted often if ever, or you might not have root permissions to edit system configurations... but you could be asked by a manager to configure your database and application to automatically launch and be fully automated by the system.
/etc and similar system directories have unique roles in the operating system. etc typically stores package and application-specific configuration files (ports, environment variables, etc.). /tmp is a very important directory for HPC, since many applications write intermediate files to /tmp if no environmental variable $TMPDIR is set in the user's profile or in the script. This directory may only have a few gigabytes allocated and is mostly meant for operating system temporary files, not intermediate files of large genomic pipelines. You may need to modify the variable to /scratch or your systems mount point for larger temporary data to be written, which may be automatically wiped out that night! Pay attention to your system administrators, since filling that directory can be catastrophic for the node if the operating system can't write small temporary files during its uptime. I'll omit a description of /mnt, /proc, /dev at this time. /usr and /usr/local may contain compiled or shared C++ object files or headers, and are generally the target during source installation (./configure && make && sudo make install), and you probably won't need to edit anything in this directory. But if you want to install software for others to use on the node, the destination is typically /usr/local/bin instead of /usr/bin or /bin, which are install locations for operating system packages and other system-wide tools or binaries.
nfs is a more advanced topic and most of us don't need to manage storage or shared access at the junior level. It&aposls worth noting however, that large genomic analyses shouldn't be run in your $HOME directory. Intermediate files should be written to the node's 'ephemeral' storage (like /scratch or /data) and final outputs or logs should be moved elsewhere before the directory is cleaned up. Don't hammer the NFS server with small mergesort files (samtools sort) or repeated writing to a log file.
Schedulers or grid-engines are part of high-performance computing clusters (cluster computing) and you might have access to one if your institution is supporting one. Your analysis or pipeline might be submitted to the grid, you might have limits for the number of simultaneous jobs, storage limits, and similar limits to be available when requesting one or more processors from one or more 'compute nodes' in the cluster. Please don't reserve multiple cores when the pipeline or tool doesn't support parallelization or you haven't configured the option in the tool (bowtie -p 8). Be aware of others you are sharing the calculation power with and plan your processing, pipelines, or parameter sweeps accordingly. If you need more power than available, talk to your advisor about cloud capabilities. I'm running 4 nodes in AWS for about $0.10 per hour. It's easier than you think!
httpd is the Apache server daemon and manages HTTP requests to different websites (static or otherwise) on a usually dedicated web server. If you want to host files or datasets, a shared drive or ftp server might be a better solution. Apache is an older and highly-customizable web server, but is often less performant for high-volume requests. nginx is a concurrent, round-robin web server that is a better bet if you're expecting higher volume of traffic (100+ sessions) for your web application.
tmux is a terminal multiplexer, similar to screen. Imagine you want to connect to a server, and your pipeline is still running when you have to leave for the night! Do you leave your laptop open and vulnerable in your lab? Do you have to awkwardly take the laptop home in your car? What if the battery runs out? What if Windows or OSX decides to automatically update and reboot your machine? What if you want to distro hop or boot up Windows for a Skype session or gaming? tmux is a session that stays open on the server you've connected to. ssh into the machine, launch a session, disconnect, launch another, disconnect, check status, disconnect, go home. When you reconnect to the session, all your windows and tabs, editors, pipeliens will still be running and in the state you left them. Check it out!
Statistics and Data Science
While we discussed briefly the spectrum of computational skills, experience, and specialization in teams above, I'll mention that data science and statistical knowledge is a main component of enterprise bioinformatics (and any other informatics position, for that matter). It's worth noting a few trends or issues that nearly every bioinformatician will face in industry.
Literate programming is becoming a strong component of condensed and reproducible research reporting and communications. Literate programming has many flavors but R-Markdown, iPython/Jupyter, and org-mode come to mind readily as efforts to make descriptions of analyses clear, concise, and reproducible. Domino is an enterprise-level reproducibility platform that merges git and AWS platforms to create highly reproducible calculations, RStudio or Jupyter sessions, and other analyses to version control datasets and deployment to cloud platforms. Without being excessively negative, I think the existence of Domino highlights a reproducibility problem within our industry, where developers leave a company, and the scripts or one-off analyses cannot be easily reproduced.
Domino can't solve the problem of poor documentation and communication, but it does make sure that any analysis run in its platform is dockerized and easy to launch in the near future. The take home from this should be that if a scientist doesn't want to read your Rmd or PDF, you can condense your language and verbosity so much, but maybe they just don't respect the quality of your research. If you don't document your code or follow software development best practices for your language or declare explicit dependencies, you probably need to look at your developer habits. If you can and often do create reports that summarize findings, literate programming is a strong tool to make reports contain the minimum amount of code to reproduce an analysis and lead other computational scientists forwards in similar modeling strategies with publication-quality graphics and document structure.
In the data munging subfield of data science, there is always a huge amount of under-structured data that we struggle to coerce into tabular formats for modeling and inference. Even in those tabular formats, sometimes considerable manipulation may be required to properly analyze subsets or groupings of the data. In the Python world Pandas, numpy/scipy, biopython, and matplotlib come to mind. In R, tidyverse tools are nearly ubiquitous. The grammar of graphics is a theoretical framework for communicating insights from data, designed for scientists to become better communicators. Don't learn it because some stupid blog told you to learn it. Learn it to the level that you feel comfortable using it. Although it is easier to use than some other graphics or stats suites, the best part about it is the theory that guides flexibility in presentation and clarity in the description of covariates, primary variables, groupings, or graphical layers(glyph, color, fill, summary stats) that reflect those categorical or numerical covariates.
As for actual modeling techniques, most scientists and bioinformaticians can do hypothesis testing and basic or intermediate regression analyses. Better foundations in these areas can have a big impact on your research. Many of those can also do clustering, unsupervised learning, and principal components analysis. Fewer still understand normalization and regularization topics to the extent that we should hope for basic and intermediate data modeling tasks. Distribution fitting is considered an advanced topic. Machine learning is a toolset for those with advanced understanding of modeling topics. It's a very strong tool and Occam's razer applies. Simple models cannot be underestimated in an age that preaches machine learning and AI are the only future of data analyses.
Just as much as we lament datasets produced from the laboratory that are under-characterized, under-sampled, or have unclear motives behind the covariate choices, it's up to us to produce clear assumptions made in calculations, better benchmarks of speed and accuracy of our tools, and rigorous null models that justify metrics or probabilities produced from our calculators. I'm sure that was the best snapple-cap advice you've ever heard of but I find the challenge so central to our field and yet often underdescribed and over-prescribed.