
Animal fats and plant oils - costs and considerations
Healthy cooking fats - mono and poly unsaturated fats - scarcity and health considerations for common animal/plant/seed fats or oils
Target audience: scientists, engineers, managers, and academic partners
Seeking a dynamic addition to your team? I bring 5 years of experience in industry biopharmaceutical R&D teams coupled with 10 years of engineering Linux systems and data analysis with Python/Perl/R/Rust. Let's elevate your projects together!

Healthy cooking fats - mono and poly unsaturated fats - scarcity and health considerations for common animal/plant/seed fats or oils

Anti-intellectualism, censorship, social media monopolies, and the wave against current Civil Rights protections

Python fundamentals guide that | s l a p s |. Read docs, help() functions, and practice, practice, prActIce!

Ubuntu + pipenv/Python3, emacs, Rstudio, pandoc, Obsidian replacements: org-mode, org-roam

An early blog post about differences between Data Analysts, Data Scientists, and Data Engineers

Short discussion on right-to-repair and technology economics for everyday Americans
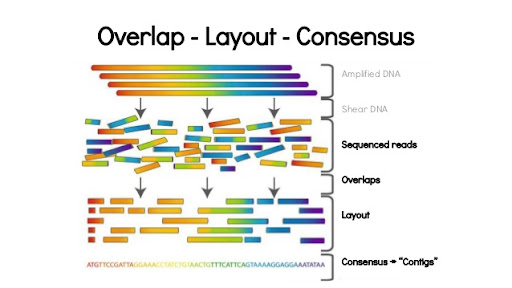
Using the networkx library for Eulerian path reconstruction and de Bruijn graphs

Reports from Civil Ministries on death tolls in armed conflicts across the world.
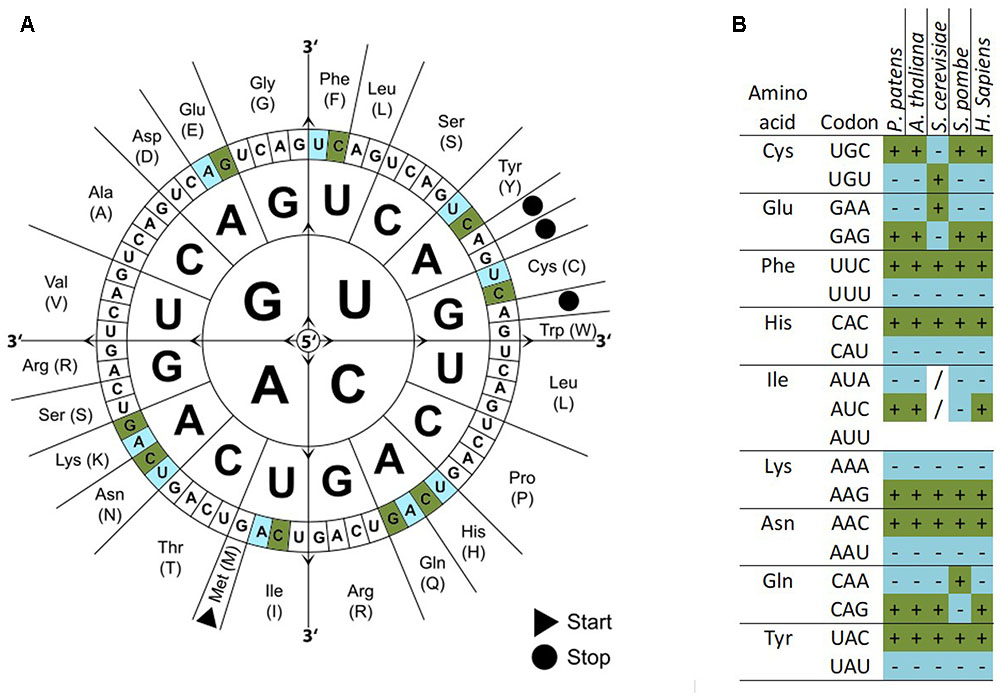
Chi-square tests, ENC, CAI, and other statistics

What are PFAS, or forever chemicals? Poly-fluorinated alkane substances (PFAS) are chemicals that have been designated as confirmed and concerning environmental toxins by governments across the gl...